Publications
Group highlights
At the end of this page, you can find the full list of publications and patents. All papers are also available on arXiv.
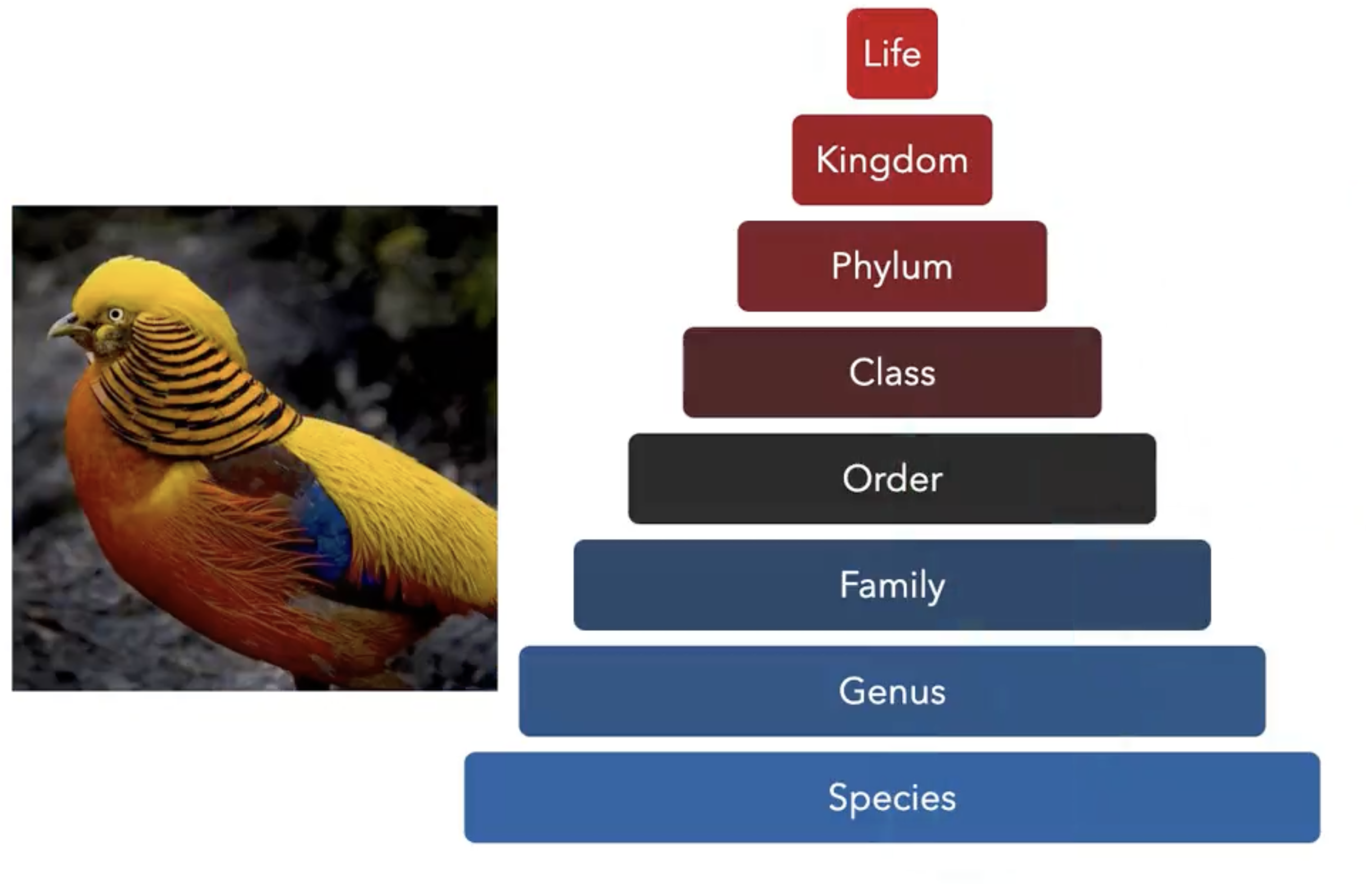
Images of the natural world, collected by a variety of cameras, from drones to individual phones, are increasingly abundant sources of biological information. There is an explosion of computational methods and tools, particularly computer vision, for extracting biologically relevant information from images for science and conservation. Yet most of these are bespoke approaches designed for a specific task and are not easily adaptable or extendable to new questions, contexts, and datasets. A vision model for general organismal biology questions on images is of timely need. To approach this, we curate and release TreeOfLife-10M, the largest and most diverse ML-ready dataset of biology images. We then develop BioCLIP, a foundation model for the tree of life, leveraging the unique properties of biology captured by TreeOfLife-10M, namely the abundance and variety of images of plants, animals, and fungi, together with the availability of rich structured biological knowledge. We rigorously benchmark our approach on diverse fine-grained biology classification tasks, and find that BioCLIP consistently and substantially outperforms existing baselines (by 17% to 20% absolute). Intrinsic evaluation reveals that BioCLIP has learned a hierarchical representation conforming to the tree of life, shedding light on its strong generalizability.
Samuel Stevens, Jiaman Wu, Matthew J Thompson, Elizabeth G Campolongo, Chan Hee Song, David Edward Carlyn, Li Dong, Wasila M Dahdul, Charles Stewart, Tanya Berger-Wolf, Wei-Lun Chao, Yu Su
See here for detailed information on dataset, model, code and demo.

In situ imageomics is a new approach to study ecological, biological and evolutionary systems wherein large image and video data sets are captured in the wild and machine learning methods are used to infer biological traits of individual organisms, animal social groups, species, and even whole ecosystems. Monitoring biological traits over large spaces and long periods of time could enable new, data-driven approaches to wildlife conservation, biodiversity, and sustainable ecosystem management. However, to accurately infer biological traits, machine learning methods for images require voluminous and high quality data. Adaptive, data-driven approaches are hamstrung by the speed at which data can be captured and processed. Camera traps and unmanned aerial vehicles (UAVs) produce voluminous data, but lose track of individuals over large areas, fail to capture social dynamics, and waste time and storage on images with poor lighting and view angles. In this vision paper, we make the case for a research agenda for in situ imageomics that depends on significant advances in autonomic and self-aware computing. Precisely, we seek autonomous data collection that manages camera angles, aircraft positioning, conflicting actions for multiple traits of interest, energy availability, and cost factors. Given the tools to detect object and identify individuals, we propose a research challenge- Which optimization model should the data collection system employ to accurately identify, characterize, and draw inferences from biological traits while respecting a budget? Using zebra and giraffe behavioral data collected over three weeks at the Mpala Research Centre in Laikipia County, Kenya, we quantify the volume and quality of data collected using existing approaches. Our proposed autonomic navigation policy for in situ imageomics collection has an F1 score of 82% compared to an expert pilot, and provides greater safety and consistency, suggesting great potential for state-of-the-art autonomic approaches if they can be scaled up to fully address the problem.
Jenna Kline, Christopher Stewart, Tanya Berger-Wolf, Michelle Ramirez, Samuel Stevens, Reshma Ramesh Babu, Namrata Banerji, Alec Sheets, Sowbaranika Balasubramaniam, Elizabeth Campolongo, Matthew Thompson, Charles V Stewart, Maksim Kholiavchenko, Daniel I Rubenstein, Nina Van Tiel, Jackson Miliko
Patents
Will be updated
Full List of publications
BioCLIP: A Vision Foundation Model for the Tree of Life
Samuel Stevens, Jiaman Wu, Matthew J Thompson, Elizabeth G Campolongo, Chan Hee Song, David Edward Carlyn, Li Dong, Wasila M Dahdul, Charles Stewart, Tanya Berger-Wolf, Wei-Lun Chao, Yu Su
ArXiv (2023)
A Framework for Autonomic Computing for In Situ Imageomics
Jenna Kline, Christopher Stewart, Tanya Berger-Wolf, Michelle Ramirez, Samuel Stevens, Reshma Ramesh Babu, Namrata Banerji, Alec Sheets, Sowbaranika Balasubramaniam, Elizabeth Campolongo, Matthew Thompson, Charles V Stewart, Maksim Kholiavchenko, Daniel I Rubenstein, Nina Van Tiel, Jackson Miliko
IEEE (2023)
You can also check publications from group members by visiting their Google Scholar page. (Click here)